Analysis on the Use of siRNA for HIV Treatment: An Investigative Report
Onyedika Moghalu – Student Researcher (Team Leader)
Alexia Joseph – Student Researcher
Duk Hyung – Student Researcher/Technical Writer
Girum Worku – Student Researcher
University of Maryland
Abstract
Current treatments for AIDS are ineffective at eradicating the virus because; they are unable to detect mutant forms of the HIV virus. At the moment, HIV AIDS has afflicted 60 million people worldwide and a staggering 16,000 new infections occur each day. The current treatment options use antiretroviral drugs consisting of protease inhibitors and reverse transcriptase inhibitors, but these current treatments are unable to permanently destroy the virus. We first performed research to understand how HIV viruses infect human cells using information obtained from sources like the Cell Digest Journal. We then investigated the current treatment drugs for HIV AIDS patients using information obtained from the Database AIDS and Cancer Research Abstract, and from molecular genetics textbooks.
We concluded that currently used drugs are ineffective at suppressing mutant viruses from replicating over a lengthy time frame. To understand the principles, the promises and, the challenges behind the use of RNAi, we investigated articles in PubMed, the Nature Journal, and the Cell Research Journal. From our findings, we concluded that although the siRNAs potential as a potent future treatment lies in its ability to directly cleave RNAs for viral genes rather than proteins, nevertheless, the technology needed to create vectors that effectively deliver the antiviral RNA to host cells has not yet been fully developed. Based on the results and conclusions from our research we cautiously recommend the use of RNAi specifically siRNA as a more effective treatment alternative for AIDS. They are effective because even when the viral genes mutates, siRNAs can be constructed to complementarily bind to specific conserved regions of the HIV’s genome. Using RNAi designed to effectively target these conserved genes means that conserved viral genes will more likely be deactivated thus, inhibiting HIV replication irrespective of its mutant form.
Key words: mutant, protease inhibitor, reverse transcriptase inhibitor, RNAi, genes, nucleotides, conserved, siRNA.
Introduction
On March 29, 2007, we received approval of our proposal to research whether the use of RNAi, specifically siRNA is a better alternative therapy for HIV treatment. This report presents the findings of our study. We proceeded to first describe how HIV virus infects the human cell and why current treatment options are ineffective at providing a cure. We researched the current treatments available and concluded that it is best to look for an alternative. We then identified and examined the principles behind the use of RNAi to inhibit viral replication, their possible limitations, and the use of siRNA in AIDS treatment.
The HIV virus arose from mutation of a similar virus called the Simian Immunodeficiency Virus (SIV), which is found in primates. It emerged in the human population of Africa in approximately 1930 (Bauman 718). The virus infects the cell by first attaching to membrane receptors of the host cell before it immediately penetrates the host cell. Once in the cell, it makes a copy of its genetic material and inserts it into the host cell DNA. Once inserted the viral DNA directs the host cell to synthesize many HIV viruses, these viruses destroy the host cell when through lysis and are released into the blood stream.
Currently, HIV has infected approximately 60 million people world wide and in fact a staggering 16,000 new infections are reported daily (Bauman 721). Several factors have significantly contributed in increasing the rate of HIV infection around the world, most notably, unprotected heterosexual contact. In addition, male homosexual contact, the use of contaminated injected needles, sharing razors and tooth brushes, and even mouth-to-mouth kissing to a lesser degree have been vital to its rapid spread.
It is estimated that infection through male homosexual contact accounted for 48% of new AIDS cases in the United States in 2002. Even with the use of condom the individual's risk of being infected is only reduced to about 70%; moreover, leading a reckless sexual lifestyle can drastically reduce the protection provided by condom use (Bauman 718).
Current treatment involves the use of highly active antiretroviral therapy (National Institute of Allergy and Infectious Disease 1-6). It contains a mixture of antiviral drugs that target the proteins produced or found in HIV virus. However, these drugs cannot eradicate the virus because the HIV genes coding for the proteins the drugs target either mutates to avoid detection, or undergo transition to a latent phase and stop producing those target proteins (Cohen par 4). As a result, HIV infected individuals currently have no hope for a possible cure. A new drug therapy is desperately needed to directly target the genes and deactivate the gene products coded for by HIV's genetic material; it is believed that use of RNAi could be very effective for this purpose (Yeung et al par 4).
Using secondary sources like the National Institute of Allergy and Infectious Disease website, microbiology and molecular genetics textbooks, and the Science Now online journal, we obtained in-depth analysis on the principles behind RNAi treatment, their efficacy and, their limitations.
We concluded that although more research on RNAi is still needed, however, compared to current treatments it seems more effective at preventing viral replication. Its ability to directly target and complementary bind to sequence specific HIV target RNA rather than viral proteins, makes siRNA better able to overcome HIV viruses that use mutant proteins to evade detection. Furthermore, siRNAs are easy to construct and test.
Despite being a better alternative to current treatments like reverse transcriptase inhibitors and, protease inhibitors, there are still challenges that need to be overcome if the full potential for this new treatment therapy is to be realized. Currently, the technology to create effective transport systems crucial to delivering the antiviral RNAs to target cells is not yet fully developed; furthermore, it is not yet known if using RNAi could inadvertently lead to the unwanted silencing of other genes vital to the cell (Rana, 2007). Nevertheless, we recommend the use of siRNA as a better alternative for HIV treatment.
In the following sections, we provide additional details about our research methods, the results we obtained, the conclusion we drew from our results, and our recommendations.
Research Methods
To better understand the mechanism of HIV infection and the use of siRNA as a treatment option, we performed the following research
- We investigated the mechanism and process of HIV infection cycle . We conduct ed an interview with an immunology expert Dr. Frauwirth to obtain current understanding of the virus and insight to future studies. We also researched recently published texts such as Kuby’s Immunology and online database from NCBI website. We were able to figure out the detailed process of HIV infection and the viral lifecycle.
- We identified the current treatment options for AIDS and their flaws: Using reverse transcriptase inhibitors . By researching the National Institute of Allergy and Infectious Disease website, we identified examples of drugs that fall under the class of reverse transcriptase inhibitors, and analyzed the biochemical pathway used by such drugs to target and prevent HIV viruses from replicating in the human cell. In addition, using information obtained from the Microbiology With Diseases by Taxonomy textbook, we identified the exact mechanism that makes reverse transcriptase inhibitors effectively attack HIV virus. Furthermore, we also researched the major flaw in using reverse transcriptase inhibitor drugs, and how the HIV virus takes advantage of this flaw to avoid being killed by drugs.
Using HIV protease inhibitors. By obtaining information on the types of drugs that fall under the class of HIV protease inhibitors using information obtained from secondary sources like Science Now Online Journal and molecular genetics textbooks, we were able to specifically analyze the reaction mechanism used by such drugs to inhibit the HIV virus from replicating. Also, we looked at the effectiveness of HIV protease inhibitors at eradicating the HIV virus and the main flaw in using such a treatment alternative. Furthermore, we examined how the HIV virus takes advantage of the limitations of using protease inhibitors to avoid being detected and destroyed.
- We researched the principles behind RNAi therapy and its use for HIV treatment. We obtained information from various journals such as the Nature Journal, and the Database for AIDS and Cancer Research, and we created a detailed outline of the pathway used during RNA interference. We also examined how such a pathway can be triggered and applied to a HIV infected cell to inhibit viral replication. We then examined the advantages and disadvantages of using RNAi.
- We determined the limitations to using RNAi. W e researched several articles from the Journal of Biotechnology and reviewed various research data from the National Institute of Health online database. In addition, I conducted an interview with an expert at the National Institute of Allergy and Infectious Diseases Dr. Frosso V oulgaropoulou to further analyze the application of RNAi to inhibiting HIV replication. We were able to identify and investigate several factors that limit the effectiveness of this technique.
Research Results
In this section, we present the results of our research. We cover the mechanism of HIV infection, the current treatment options, the principle behind microRNA therapy, and the limitations of microRNA as a drug for HIV treatment. The following results correspond to the tasks described in the research methods section.
1. The process of HIV infection
Primary binding of HIV. HIV binds to specific cells in the body that express certain surface protein. The specific cells include T-cells and monocytes, which serves very important immunological roles. The primary binding of HIV occurs between CD4 and gp120. CD4 is specific surface protein that is expressed in both T-cells and monocytes, and gp120 is specific surface glycoprotein on HIV that can bind to CD4 (Frauwirth, Interview) .
Secondary binding of HIV. The primary binding between gp120 and CD4 induce secondary binding. The secondary binding occurs differently in T-cells and monocytes. T-cells have specific receptor called CCR5 on the surface, and on the other hand monocytes have another receptor called CXCR4. These two specific receptors can bind to gp41, glycoprotein embedded in the membrane connected to gp120 (Frauwirth, Interview) .
Fusion of host and HIV. The primary and secondary binding of HIV induces fusion of HIV membrane with host membrane. The fusion is mediated mostly by secondary binding causing the fusion mechanism, but primary binding is necessary for secondary binding to occur. The fusion mechanism is triggered when the CCR5 or CXCR4 is activated by gp41 and lead to serious of events in the host to allow HIV to fuse (Frauwirth, Interview) .
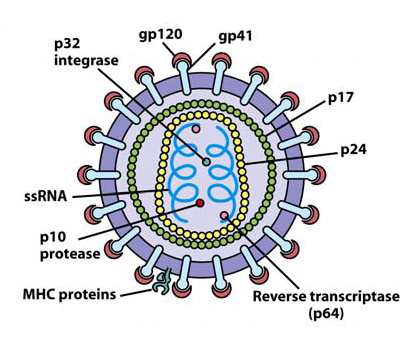
Insertion of HIV genome into the host genome. The fusion of host and HIV membrane releases the viral genome and viral proteins into the host. The most important viral protein called reverse transcriptase can make viral RNA into RNA-DNA hybrids, and converted to normal double stranded DNA. Double stranded DNA is required to infect the host, because in order to reproduce and expend, it require the host to make viral protein and more copies of viral RNA, and only double stranded DNA is capable to fulfill the task. After producing double stranded DNA, another type of viral protein called integrase insert the viral double stranded DNA into the host genome at random location and become provirus (Kuby et al 2007) . The figure 2 illustrates all the components described and indicate the anatomical locations of the components to clarify some of the explanation. The term provirus refers to the state of virus infection, which has no significant activity in the host.
Fig. 1: The HIV viral structure
Source: T homas K. Kuby. “Immunology.” New York: W.H. Freeman and Company; 2007 Fig. 20.9aActivation of provirus and reproduction. The provirus gets activated when the environmental or internal signal cause the transcription factors stimulate transcription of proviral DNA. The transcription factors promote the process that allows DNA to pass the contained information to produce messenger RNA, which can ultimately produce proteins. The whole process is done by using host cellular machinery (Frauwirth, Interview) . The final outcome of this process is mass production of viral proteins and viral RNA. The products then assembles itself to form immature HIV, then buds out from the host and released HIV complete maturation and infect other cells. The term bud refers to the process of which HIV uses the host membrane containing all the necessary viral proteins such as gp120 and gp41 to form viral membrane and separate itself from the host (Kuby et al 2007) .
2. The Current AIDS Treatment Options:
We studied two main options for treatment:
Treating AIDS using reverse transcriptase inhibitors drugs . There are two subclasses of drugs that fall under reverse transcriptase inhibitors, nucleotide reverse transcriptase analogs, and non-nucleotide reverse transcriptase inhibitors (Bauman, 2007).
Reverse transcriptase inhibitors act by interfering with a critical step in the HIV viral cycle called reverse transcription. During reverse transcription, an enzyme called reverse transcriptase converts HIV RNA (ribonucleic acid) to HIV DNA (deoxyribonucleic acid). Examples of drugs that are nucleotide reverse transcriptase inhibitors are AZT (Azidothymidine), Retrovir and Zidovudine.
a) Nucleotide reverse transcriptase inhibitors drugs. These drugs are made up of primarily faulty nucleotide (DNA) sequences, when these faulty DNA sequences are embedded into the newly synthesized viral DNA by the reverse transcriptase enzyme, synthesis of the nascent viral DNA is stopped thus preventing the virus from completely replicating its genes (National Institute of Allergy and Infectious Disease, 2006).
b) Non-nucleotide reverse transcriptase inhibitors . These types of drugs can be proteins that attach directly to the active site of the reverse transcriptase enzyme. When bound to the active site, the inhibitor induces a conformational change in the reverse transcriptase that prevents the enzyme from being able to convert the HIV RNA to DNA. Without the HIV’s genetic information being in a DNA format it can’t be integrated into the host cell’s chromosome, thus, the virus cannot direct the human cells to synthesize HIV virions, therefore the virus is unable to replicate. An example of a non-nucleotide reverse transcriptase inhibitor drug is Nevirapine (Hartwell et al., 2004).
Although reverse transcriptase inhibitors effectively inhibit the HIV cycle of growth and replication, nevertheless, these drugs are unable to cure AIDS because they do not completely eliminate the HIV virus from the body (Cohen, 2002). For the nucleotide reverse transcriptase inhibitor drugs, their major flaw comes from the fact that when administered to the patient at high doses it is extremely toxic. As a result, doctors only administer the drugs at low doses that do not destroy all the viral particles (National Institute of Allergy and Infectious Disease, 2006).
For non-nucleotide reverse transcriptase inhibitor drugs, their major flaw is that they are unable to keep up with the high mutation rate of the HIV virus. Because the viral genes coding for the reverse transcriptase enzyme mutates frequently, the HIV virus can produce alternate forms of the same enzyme that are structurally altered in such a way that their active sites cannot be recognized and attached to the inhibitor drugs. Consequently, the non-nucleotide reverse transcriptase inhibitor drugs cannot identify and inactivate these mutant enzymes and so the HIV virus can go on replicating even in the presence of the antiviral drugs.Treating AIDS using protease inhibitor drugs. These inhibitors act by interfering with the protease enzyme that the HIV uses to produce infectious viral particles, examples of protease inhibitor drugs include, Norvir, Crixivan, Viracept and, Lexiva. During HIV replication, the virus produces two inactive proteins called gag and pol proteins, these proteins need to be cleaved in order to become activated. The cleavage of gag by HIV protease generates the viral coat used by HIV to attach to the human cells, and cleavage of pol generates key HIV components such as reverse transcriptase, capsid, and matrix proteins. The HIV protease inhibitor drugs disrupt HIV viral replication by binding to inactive gag and blocking its active site so that it cannot be activated by protease (Bauman, 2007). Consequently, the inactive gag is unable to generate viral components needed to form a truly functional virus as a result, HIV virus is unable to produce viable new viruses; therefore, it cannot replicate and destroy the human cells.
The major flaw of protease inhibitors results from their inability to keep up with the high rate of mutation of the HIV genes encoding the protease enzymes (Cohen, 2002). As a result, the virus is able to synthesize so many variant forms of the protease enzymes each having slightly altered active sites that are unrecognizable to the protease inhibitors. When drugs like Norvir are administered to patients they are unable to attach to these modified active sites and deactivate the mutant gag proteins (Hartwell et al., 2004), consequently the virus is able to successfully multiply within the patient even though they are on a rigorous treatment regimen.
3. Principles behind RNAi and its therapeutic purposes:
RNAi (RNA interference) is basically the process of inhibiting a genes expression by using double stranded RNA to cleave the genes RNA transcript or preventing the RNA from being translated to a protein. RNAi is quite ubiquitous in nature and can be found in fungi, animals and, plants (Engels and Hutvagner, 2006). The process of using double stranded RNA was first observed in worms (Caenorhabditis elegans). In plant cells, RNA interference is used to inhibit adenoviral attack; it is a tool used by plants analogous to an animal’s use of immune cells to fight viral infections (Yeung, et.al 2005).
For our research analysis we describe the two major RNAi mechanisms: the use of siRNA (small interference RNA) and, miRNA (microRNA). siRNA are 21 to 23 nucleotides long double stranded RNA that deactivates a target messenger RNA (mRNA) by binding and directly cleaving it (Strachan and Read, 2004). To understand the efficacy of siRNA and the RNAi pathway it induces, it is imperative we first provide a description of how siRNA is generated.
A small interfering RNA as shown in Appendix C originates from a linear double stranded RNA (dsRNA). This double stranded RNA is made up of two strands: an antisense strand (guide strand) and a sense strand (passenger strand). The antisense strand contains nucleotide sequences complementary to the target RNA, while the sense strand contains sequences complementary to the guide strand. Once the linear double stranded RNA is formed, it binds to an RNAse II enzyme called DICER (Rana, 2007). The DICER cleaves the dsRNA into a 21 to 23 nucleotide long RNA known as siRNA. Now that the siRNA has been formed, we proceed to examine how it triggers gene silencing. siRNA acts by complementarily binding to a complex called RISC (RNA-Inducing Silencing Complex) through its guide strand to form an siRNA-RISC complex. This complex now binds specifically to the target RNA and cleaves the target RNA thus, preventing the gene product coded by the RNA to be produced, as shown in Appendix C.
Fig 2: m icroRNA active strands
Source: http://www.ma.uni-heidelberg.de/apps/zmf/argonaute/interface/mirna.jpegMicroRNA on the other hand is generated from a markedly different process. miRNA as seen in figure 2 are 22 nucleotide long RNA molecules coded for within the genome that lead to gene silencing (Strachan and Read, 2004). Unlike siRNA, microRNAs are actually coded for by genes in the cell. The miRNA genes are transcribed by RNA polymerase II in the nucleus, to form a primary miRNA transcript. Just like normal messenger RNAs, the primary miRNA formed has a 5’ cap and a poly-A-tail. Like siRNA, it is double stranded with guide and passenger strands, in addition, it is also looped (Rana, 2007).
The primary miRNA are shortened by a nuclear RNaseII called DROSHA into what is called a premiRNA. The premiRNA are then carried from the nucleus of the cell to the cytoplasm. While in the cytoplasm the premiRNA is cleaved by DICER into the 22 nucleotide mature microRNA. Once formed the microRNA now initiates its RNA interference pathway. It does so by binding the RISC via its guide strand to form a complex. It is this complex that binds to the target RNA albeit with a lower degree of complementarity.
A current view is that miRNAs function to silence gene expression through imperfect base-pairing with transcripts. Due to this imperfect binding by the guide strand, it does not cleave the RNA but rather the loop created as a result prevents translation from occurring. Since RNA silencing mediated by miRNA does not require complete sequence complementarity, one miRNA can target multiply different miRNAs (Engels and Hutvagner, 2006). Thus, miRNA prevents the gene product coded for by the target RNA from forming (Rana, 2007).
Based on our research results from the National Institute of Allergy and Infectious Disease and also from the Nature Journal, current findings support such a notion since several viruses have been found to encode RNAi suppressors, which could function to influence the cell's overall miRNA environment. RNAi, apart from providing an indispensable reverse genetic tool and promising therapeutic approach, regulates gene expression on both transcriptional and post-transcriptional level and it is used to fight against viruses and bacteria (Engels and Hutvagner, 2006).
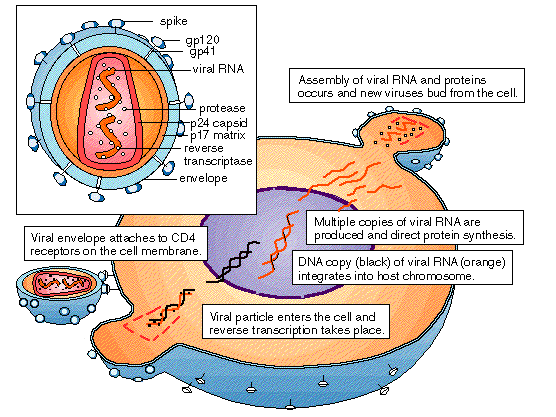
Now that we have an idea of the gene silencing pathways used by both siRNA and microRNA, let us see how our knowledge of the RNAi process can be specifically applied to inhibit HIV viral replication using siRNA. Studies on the use of siRNA to inhibit HIV-1 replication have shown promise. As seen in figure 3 the HIV viral core consists of several proteins such as reverse transcriptase, capsid and, protease. Scientists have used siRNAs to specifically target RNA transcripts coding for key HIV-1 protein components such as tat protein, reverse transcriptase, and the HIV-1 CD4 co-receptors. Although such studies showed that siRNA was able to significantly suppress HIV-1, nevertheless certain issues remain of concern (Rana, 2007).
NOTE : HIV is a retrovirus with an outer envelope containing proteins, some of which are spikes consisting of gp20 and gp41 proteins. The core of the virus contains RNA genes and copies of reverse transcriptase, the enzyme that makes a DNA copy of RNA. During the reproductive cycle, the DNA copy first becomes integrated into a host chromosome and then is transcribed back into viral RNA.
Fig. 3: Core of HIV retrovirus Source: http://www.mhhe.com/biosci/genbio/maderbiolog
One of the major concerns about the use of siRNA based therapy is the question of how to effectively deliver the siRNA to the target cells without it being degraded. Recent approaches aimed at tackling this question have not only yielded novel strategies for RNA delivery to cells in vivo, but they have also provided fantastic new strategies to suppress HIV-1. Among strategies used to deliver siRNA to HIV-1 infected cells is the use of lentiviral vectors, and also RNA packaged in micelles (Morris and Rossi, 2006). We found the use of certain lentiviral vectors to be absolutely fascinating especially when you consider how it could also contribute to preventing HIV virions from being produced and released from the infected cell.
For instance, let us consider the use of conditionally replicating HIV vectors (crHIV vectors). In crHIV vectors the antiviral siRNA in addition to the vector RNA are packaged together, unlike a normal HIV virus the crHIV vector lacks the gene and protein for the tat and other structural polyproteins necessary for the formation of nascient viruses in the host cell. Once created, the crHIV vector is targeted to an already infected HIV cell. Like a normal HIV virus it penetrates the host cell and releases its vector RNA along with the antiviral siRNA. The vector RNA is converted to DNA and integrated into the infected cells genome.
However, because the vector RNA lacks the gene for the structural polyproteins necessary for the production of its vector virions, it competes for the tat protein used by the HIV-1 virus already in the host cell for replication. In the mean time, the antiviral RNA introduced into the host cell targets the HIV-1 RNA and cleaves it, consequently reducing the amount of HIV-1 virions that are produced and thus fewer “demand” for the tat polyprotein by the HIV-1(Morris and Rossi, 2006).
As a result, the crHIV vector gains the advantage and out competes the HIV-1 virus for the tat protein therefore the vector virions are predominantly produced and released from the infected cell compared to HIV-1 virions. The use of RNAi to inhibit HIV-1 replication provides significant advantages that make them a very appealing therapeutic strategy. RNA interference using siRNA are easy to construct and test in cells, furthermore, they cost less to create and it takes them less time to elicit their antiviral effects against the HIV-1 virus (Morris and Rossi, 2006).
One of the current major obstacles in the treatment of AIDS has been the fact that the HIV virus is able to evade antiviral drugs such as protease and reverse transcriptase inhibitors through varied mutations on their respective genes (Cassol et al, 2006). The virus also avoids being destroyed by using alternative splicing to try to avoid being targeted via RNAi. To try to overcome this problem, scientists are currently attempting to create siRNAs targeting conserved regions such as the promoters of the HIV-1 that are much less susceptible to mutations. The principle to this strategy is that because the fidelity of such regions of the viral RNA is absolutely crucial to the virus’s ability to code information needed for replication, the HIV-1 virus ensures that very little mutation is permitted in those regions (Morris and Rossi, 2006).
4. Limitations to RNAi Therapy Using siRNA
From our research, we found three main limitations of the microRNA techniques in treating HIV: Although RNAi therapy holds great promise nevertheless, like all other drug therapies for HIV treatment it does have some limitations.
Potential for off targeted gene silencing, siRNAs constructed to cleave RNA for a specific gene could lead to the silencing of genes not meant to be targeted. The effect of such inadvertent silencing could lead to unforeseen effects in the host organism (Morris and Rossi, 2006).
Inefficient RNA delivery to host cells. Scientists have made great strides in creating systems especially in the case of HIV treatment, capable of carrying the siRNA in the blood to the target cell. One of the key strategies employed is the use of lentiviral vectors packaged with the antiviral siRNA and delivered to the host cell. As mentioned earlier, the use of conditionally replicating HIV vectors have also been used. However, efficient delivery systems are still sought after in the treatment of other viral-related diseases.
Viral Resistance via Mutations. Point mutations and alternate splicing by the virus within the regions targeted by the siRNA can lead to the virus developing nucleotide sequences that are not specific to the siRNA. Thus, the virus might develop resistance to the therapeutic strategy. (Morris and Rossi, 2006).
Conclusions
The conducted interview with Dr. Kenneth Frauwirth and subsequent research revealed several unique characteristics of HIV infection. The results described the process of infection, reproduction and involved proteins that might be potential target for treatment. The first unique characteristic was binding of HIV to host. The host surface protein CD4 serves unique and important roles in host, but CCR5 and CXCR4 has less important role in the host, which indicate blockage of CCR5 and CXCR4 can reduce the infection by HIV. Another unique characteristic is reverse transcriptase, which is only found on the sub class of virus called retrovirus, which include HIV. It is currently the most used target for the HIV treatment, but still needs improvement. The process of HIV infection reveals more characteristics that can be used as target for treatment.
The major current treatment options of HIV infected patients involve the use of two major classes of drugs, reverse transcriptase inhibitors, and HIV protease inhibitors. When these treatment options are used they are initially successful at inhibiting viral replication. However, the HIV virus eventually develops resistance by altering its genes and changing the structure of the proteins targeted by these drugs so that they won’t be recognized. Consequently, we conclude that both current treatment options are not effective at totally stopping the HIV virus from replicating within the human body.
The use of RNA interference especially via siRNA to inhibit HIV replication is based on the idea of targeting viral RNA with complementary RNA that ultimately inhibits the viral gene product from being made. Using siRNA based therapy for AIDS treatment holds certain advantages that make it a promising treatment option. siRNAs are easy to construct and relatively cost efficient. They can be packaged in lentiviral vectors and delivered into the target cell. In addition, the use of siRNA unlike protease inhibitor drugs can employ strategies designed to overcome a key obstacle to the efficacy of protease and reverse transcriptase inhibitor drugs. To overcome the resistance inferred by HIV-1 viruses through their ability to mutate target sites, scientists believe siRNAs can be constructed that target conserved regions such as, promoter regions. A key disadvantage to using RNAi as a treatment is the possibility of inadvertent off targeted gene silencing in vivo.
Ultimately, even though siRNA treatment seems to be an effective tool to fighting the HIV-1 virus it is likely to be used in the future in conjunction with other treatment options to create a very powerful highly active antiretroviral treatment cocktail.
Recommendations
Current research shows that people with mutations on CCR5 and CXCR4 receptors allowed the HIV positive patient to live without getting AIDS. These mutations alter the receptor binding mechanism thereby prevent HIV from entering the host. Due to the inability of reverse transcriptase and protease inhibitor drugs to totally destroy the HIV virus and stop the progression of AIDS, it is possible that RNA interference pathways triggered via siRNA can be used to cleave the host RNA coding for these receptors in stem cells for primary T cell lines, thereby preventing expression of the genes for these receptors. Information obtained from ongoing and future experimental studies using RNAi will give us a much greater insight on how to utilize this rapidly developing genetic tool to effectively fight viral related diseases such as AIDS. Until we find ways to resolve some of the risks associated with RNAi use especially with siRNA, it is best to remain cautious rather than overly optimistic in our assessment of this therapeutic approach.
References
All The Virology On The WWW. Ed. David M. Sander. 2007. http://www.tulane.edu/~dmsander/ATVdict.html
Ambion The RNA Company. 2007. 23 July 2007. http://www.ambion.com/techlib/resources/miRNA/mirna_intro.html
Bauman, Robert. Microbiology With Disease by Taxonomy. 2 nd ed. Pearson: Benjamin Cummings, 2007.
BM Engels and G Hutvagner, “Principles and effects of microRNA-mediated post-transcriptional gene regulation.” Nature Journal. Oncogene (2006) 25, 6163–6169
Cassol, E., Alfano, M., Biswas, P., Poli, G.,” Monocyte-derived macrophages and myeloid cell lines as targets of HIV-1 replication and persistence.” 2006, J. Leukoc. Biol. 80: 1018-1030
Cohen, Jon. “Why Drugs Can’t Keep HIV Down” Science Now 7 Aug.2002. 19
Frauwirth, Kenneth. Interview. 2 May . 2007
Hartwell, H., Hood, L., Goldberg, M., Reynolds, A., Silver, L., & Veres, R. (2004). Genetics: From Genes To Genomes. New York: McGraw- Hill.
Morris, K.V. and Rossi, J.J. “Lentivirus- Mediated RNA Interference Therapy For Human
Immunodeficiency Virus Type 1 Infection”. Human Gene Therapy. 17 (2006) 479-486.
Omoto S, Fuji YR, “Regulation of human immunodeficiency virus 1 transcription by nef microRNA.” Journal of Gen Virol. 2005; 86:751–755. doi: 10.1099/vir.0.80449-0. [ PubMed]
Rana, M. Tariq.” Illuminating the silence: understanding the structure and function of small RNAs.” Nature Reviews. Molecular Cell Biology 8, (January 2007): 23-36.
Sigma-Aldrich. 2007. 23 July 2007. http://www.sigmaaldrich.com/Area_of_Interest/Life_Science/Functional_Genomics_and_RNAi/Key_resources/RNAi_Glossary.html
Strachan, Tom., and Read P. Andrew.” Genetic Manipulation of Cells and animals”. Human Molecular Genetics 3. Ed. Fran Kingston and Michael Morales. Garland Publishing, 2004. 592-595.
Talking Glossary. Ed. Judith Wyatt. 2007. http://www.genome.gov/glossary.cfm
Thomas K. Kuby. “Immunology.” New York: W.H. Freeman and Company; 2007
Treatment of HIV Infection.18 Aug 2006. National Institute of Allergy and Infectious Disease. 19 Mar.2006. http:// www.niaid.nih.gov/factsheets/treat_hiv.htm
Voulgaropoulou , Frosso. Interview. 7 August . 2007
Yeung Man Lung, “Changes in microRNA expression profiles in HIV-transfected human cells.” BioMed Central Ltd. 2005 December 28. doi: 10.1186/1742-4690-2-81.
Zhou, M., Deng., “Coordination of Transcription Factor Phosphorylation and Histone Methylation by the P-TEFb Kinase during Human Immunodeficiency Virus Type 1Transcription” (2004).. J. Virol. 78: 13522-13533
Glossary
Conserved Sequences: A DNA sequence that is very similar in different species or organisms. It has very few mutations.
DICER: A member of RNase III class of ribonucleases used in the RNA interference pathway to cleave double stranded RNA to 21 base pairs.
DROSHA : A nuclear RNaseII used to shorten primary miRNA into what is called a premiRNA
Gag: Lentiviral packaging element that codes for a structural protein.
Genes: Region of DNA coding information used to form either a functional RNA or an actual protein.
microRNA: Small RNA molecules encoded in genomes of plants and animals. They regulate gene expression by binding to 3’ untranslated region of the target RNA.
mRNA: The RNA produced directly from DNA transcription. It carries the transcribed information from the nucleus to the cytoplasm, to be used for translation.
Mutation: A change in DNA sequence between generations.
Nucleotides: Monomers of nucleic acids. They are made of ribose or deoxyribose, phosphate groups, nitrogenous bases.
Pol: Lentiviral packaging element that codes for proteins.
Reverse Transcriptase: Enzyme that converts RNA to DNA.
RISC: An siRNA directed endonuclease that catalyzes the cleavage of a single phosphodiester bond on the mRNA target.
RNAi: A technique where double stranded RNA slice specific RNA transcripts.
siRNA: 21 – 23 nucleotide double stranded RNA molecules that cause the cleavage and degrading of a target RNA.
Tat: A lentiviral element needed for the elongation of nascent viral transcripts.
Vector: A molecule used to propagate a nucleic acid or even a protein.
Virion: A mature virus.
List of Abbreviations
Abbreviation
Meaning
AIDS
Acquired Immunodeficiency Syndrome
AZT
Azidothymidine
DNA
Deoxyribonucleic Acid
HIV
Human Immunodeficiency Virus
miRNA
MicroRNA
nt
Nucleotides
RISC
RNA Induced Silencing Complex
RNA
Ribonucleic Acid
RNAi
RNA interference
siRNA
Small Interfering RNA
APPENDIX A : Interview with Prof. Kenneth Frauwirth
The following interview took place on May 2, 2007. A transcript of the interview follows.
(D) – Duk Hyun, Interviewer
(K) – Kenneth Frauwirth, PhD. Expert Immunologist
D – Thank you Dr. Frauwirth for agreeing to have an interview with me today.
K – It is my pleasure to inform students on their curiosity in the field.
D – So, for the first question. Could you explain the infection mechanism of HIV?
K – As you know HIV binds to CD4+ T- lymphocytes and monocytes through CD4+ and gp120 interaction. This binding cause the confirmation change in gp120 to allow gp41 to bind to certain chemokine receptors such as CCR5 on T-cells and CXCR4 on monocytes, and this binding cause G-protein linked cascade and ultimately fusion occurs between HIV and host membrane.
D – Then is it possible to block the second interaction to prevent HIV from infecting the host?
K – It is not entirely sure, but the current studies show that complete state of latency in HIV+ patience with mutations in those receptors, so it is certainly possible approach for treatment.
D – One more question, so after the infection and integration of viral genome. What happens to the inserted viral gene?
K – The state of viral gene in host genome is called provirus stage and it is not entirely sure if it is in state of latency or has minimal activity. But after certain stress triggers the transcription factor to start transcription which is much rapid than regular gene transcription due to LTR on both side of viral gene. Then it follows the regular gene expression mechanism to form viral RNA and viral proteins using host machinery.
D – Thanks again for the interview Dr. Frauwirth. I’ve learned much from the interview.
K – You’re welcome. If you have any more questions feel free to email me.
APPENDIX B : Interview with Dr. Frosso Voulgaropoulou
This interview took place on August 7, 2007. A transcript of the interview follows.
(O) – Onyedika Moghalu, Interviewer
(F) – Frosso Voulgaropoulou, PhD. National Institute of Allergy and Infectious Disease.
O – Thank you Dr. Frosso for agreeing to have an interview with me today.
F – No problem at all, I am quite impressed that you chose to research on such a complex topic.
O – First let me clarify my understanding of siRNA, miRNA and how it is applied to HIV infection. siRNAs are basically 21 to 23 nt long RNA sequences that bind with high level of complementarity to a target RNA strand. They prevent a gene from being expressed by directing enzymes that cleave the target strand. microRNAs on the other hand are 22-23nt long sequences coded by genes in the cell. microRNAs act by preventing translation rather then cleaving the target RNA.
F – Exactly, microRNAs are broad acting; they don’t have to be perfectly complementary to the target RNA unlike siRNAs that need to be almost perfect. Thus, microRNAs can lead to inadvertent gene silencing due to their lack of specificity. As a result, they would not be the ideal therapeutic tool in fighting HIV replication in a cell. Unlike siRNAs that actually cleave the mRNA, microRNAs form loops due to non-complementary base pairing, and these loops prevent the proteins needed for translation from being able to properly attach to the mRNA.
O – I was reading the article on RNA silencing you gave me, and it says a single mutation between the antisense strand and the RNA transcript drastically reduces the ability of siRNA to target genes.
F – Yes, otherwise it won’t cleave the RNA transcript. Remember when you took biochemistry and you learned about how proteins have primary, secondary, tertiary and, quaternary structures. The same applies to the principles behind the use of siRNA to cleave a target. The siRNA- directed enzymes that cleave the RNA transcript need to be in a specific secondary conformation in order to be able to act on their target. A single mutation in the anti sense strand affects this secondary structure. When you have non perfect complementarity between the target and the siRNA, the siRNA can behave as microRNA by forming a loop and triggering the microRNA interference pathway.
O – In a molecular genetics class I took, we talked about the idea of methylation of genes in order to make them inaccessible. Is targeted methylation possible? Can it be induced in receptors such as CD4 receptors targeted by HIV so that they won’t be expressed. (Use of RNA dependent DNA methylation)?
F – Targeted methylation is possible but it is an extremely difficult approach. Methylation is a mechanism that an organism uses to prevent gene accessibility. Applying directed methylation in HIV infected cells can lead to unforeseen inadvertent physiological effects. I am not that familiar with the use of methylation to silence genes.
O – siRNA resistant viruses can come about by single nucleotide changes.
Must the region targeted by siRNA be approximately 21 to 25 nucleotides long? If not, then why not build much shorter siRNA strands that target shorter regions on the target RNA. By doing so, the smaller the target region the less alternate mutant forms that the virus can employ within that region.
F – First let us consider what happens if the siRNA is too long. If their size were too big they could cause the body to trigger its interferon response. Thus, all the host cells containing siRNAs are recognized and destroyed and this would be catastrophic for the organism. Amazingly, it is thought that RNAi evolved in congruence with the body’s immune system so that their length would be such that they would not trigger an immune response. If the siRNAs were too small it will make it very difficult to properly anneal to the target RNA transcript. Thus, the siRNAs would only weakly bind to the target and would not produce sufficient interaction needed to activate cleavage due to random interaction.
O – All HIV viruses no matter how variant still share certain fundamental physical (structural and functional protein) characteristics. Thus, they all have conserved genes. Since these genes are probably least likely to mutate why not target siRNAs to them.
F – Your idea makes perfect sense. But the problem with these conserved regions is that they are located inside the virus (beyond the viral coats). Because these genes are encapsulated in the virus and surrounded by a highly variable viral coat, it is difficult to attach and penetrate the viral coat to get at these conserved regions due to the fact that the genes coding for the receptors on the viral surface are constantly changing. The only time that these conserved regions can be freely accessible is when the viral genes have been released inside the host cell during infection.
As a side note, when the HIV virus develops resistance to drugs their replication slows down. The state of a virus in the host cell in response to treatment is determined by its cost fitness. This is the ability of a virus to successfully replicate and be virulent. Each time the virus mutates in response to the treatment regimen, its replication rate slows down while its virulence increases. A good strategy to treating AIDS might be to target many of its different conserved genes thus, making it extremely difficult for the virus to mutate at all these target sites simultaneously. Yet, HIV still manages to evade targeted treatment.
O – Have any field tests been done on animals, humans?
F – Not yet, although a clinical trial is about to be underway!
O – Thank you so much for sharing your time to answering my questions. I am taking a virology course next semester and I hope to apply the knowledge gained from the course into better understanding how HIV replication can be more effectively controlled.
F – It has been my pleasure. Keep in touch if you have any more questions.
APPENDIX C : siRNA synthetic and interference pathway
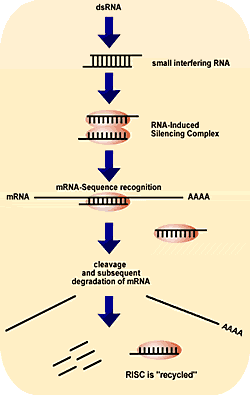
Source: http://www.ohsu.edu/research/core/images/sirna_MWG-biotech.gif
|